Indica Labs Advance Molecular Pathology with Multiplex RNA FISH Quantification Software
Indica Labs Advance Molecular Pathology with Multiplex RNA FISH Quantification Software
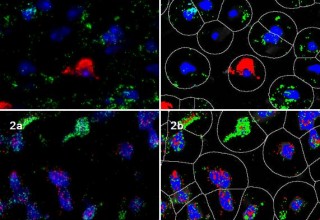
Online, January 2, 2013 (Newswire.com) - Indica Labs, Inc. announced today the release of a revolutionary molecular pathology software module for automated analysis of fluorescent RNA In situ Hybridization (RNA ISH) assays. This latest software application is the first of its kind, offering whole-slide quantification of multiplexed RNA biomarkers on a cell-by-cell basis. The software rapidly generates single-cell RNA expression profiles for hundreds of thousands of cells in microscopic fluorescent images, allowing researchers to precisely measure overall expression ratios and cellular heterogeneity across entire tissue sections, tissue microarrays, and other microscope slide-based applications.
Molecular RNA assays have moved increasingly to the mainstream for major biomedical research areas including oncology and neuroscience. More recently, advancements have been made by the introduction of chromogenic RNA ISH for brightfield interpretation and multiplex RNA FISH for measuring multiple RNA biomarkers simultaneously. Until now there has been a lack of software for true cell-based quantification of these new multiplex assays. Indica has developed the Multiplex RNA FISH software for assays like RNAscope®, an award-winning multiplex single-molecule RNA ISH technology developed by Advanced Cell Diagnostics (Hayward, California). ACD CEO Yuling Luo, PhD. remarked "Multiplex quantitative analysis of gene expression in single cells while preserving cellular context is very important in today's biomedical research and molecular diagnostics. It should greatly facilitate the study of cellular heterogeneity that underlies nearly all of the biological and disease processes".
Indica's Multiplex RNA FISH module combined with their Chromogenic RNA ISH module and HALO™ platform have propelled Indica Labs' leadership with the most advanced software technologies for molecular pathology image analysis. Indica CEO, Steven Hashagen commented "With the release of this software, Indica Labs offers the most complete RNA ISH quantification suite for both brightfield and fluorescence; meanwhile we have a number of exciting related technologies in development."
For more information please visit http://indicalab.com/product-fluorescent-rna-ish.html or contact info@indicalab.com.